Experimental Thoughts 3: Technical Implementation
Posted on Sun 01 June 2025 in Thoughts
From the last two articles, we know we need a tool to help our experiments. This began by asking what we demand from our experiments. Now, I describe how the tool mitosis provides those benefits and more.
Let's collect thoughts
To recap, we want experiments to be:
- Reproducibile
- Ran in a separate process, from start to finish, with dependency list, environment variables, and git information automatically recorded.
- Reusable
- An experiment is an importable Callable, written with important factors exposed as parameters and metrics as return types. Individual runs, or trials, result in artifacts separate from the experiment definition.
- Describable
- A dictionary is used to name arguments or groups of arguments. This dictionary enforces a single source of truth, so that different trials using the same argument name must result in object equality. Should be a way to distinguish parameters that matter and those that don't.
- Composable
- Some experiments should be thought of experiment steps, providing their data as well as intermediate metrics. The experiment runner needs to parse their output and forward the data to the correct parameter of the next step.
Some challenges spring to mind. The kinds of equality checks across processes in reproducible and describable require some sort of serialization to disk, as well as either deserialization or a correspondence between serialization equality and object equality. Secondly, multiple processes querying the disk will imply some sort of distributed systems challenges.
Other important wishes include producing plots for a trial. It also might be nice to have an experiment browser. Finally, we'll note that running the experiment is likely the dominant cost; Almost no plotting, or even interaction with the disk, will take as long as a large matrix inversion. Thus, we'll consider the admin work of an experiment runner as effectively free.
The experiment runner
We have stripped several parts of what some consider an experiment into a project-independent experiment runner, yet other project-specific parts, like the argument definition dictionary, remain unhomed.
Project layout
At this point, let me share the project organization that works for me.
- A package with the method under investigation.
- A package of experiments, exposed as a library of functions. Some methods are wrapped in local sklearn.BaseEstimator to support the dichotomoy of of data-independent initialization and data-dependent fitting.
- A project-independent runner (mitosis)
- An overall repository configured with what experiments to investigate, a dictionary of plain english names for arguments to investigate, a script file of all the experiments to run, and a directory for mitosis to write experiment artifacts. It also sometimes has the LaTeX files for writing a paper, as well as a post package for generating final images for papers/talks from experiment records. For the especially savvy, post has a module varaible for saving files in the folder where LaTeX will
A graphical example of the packages is below:
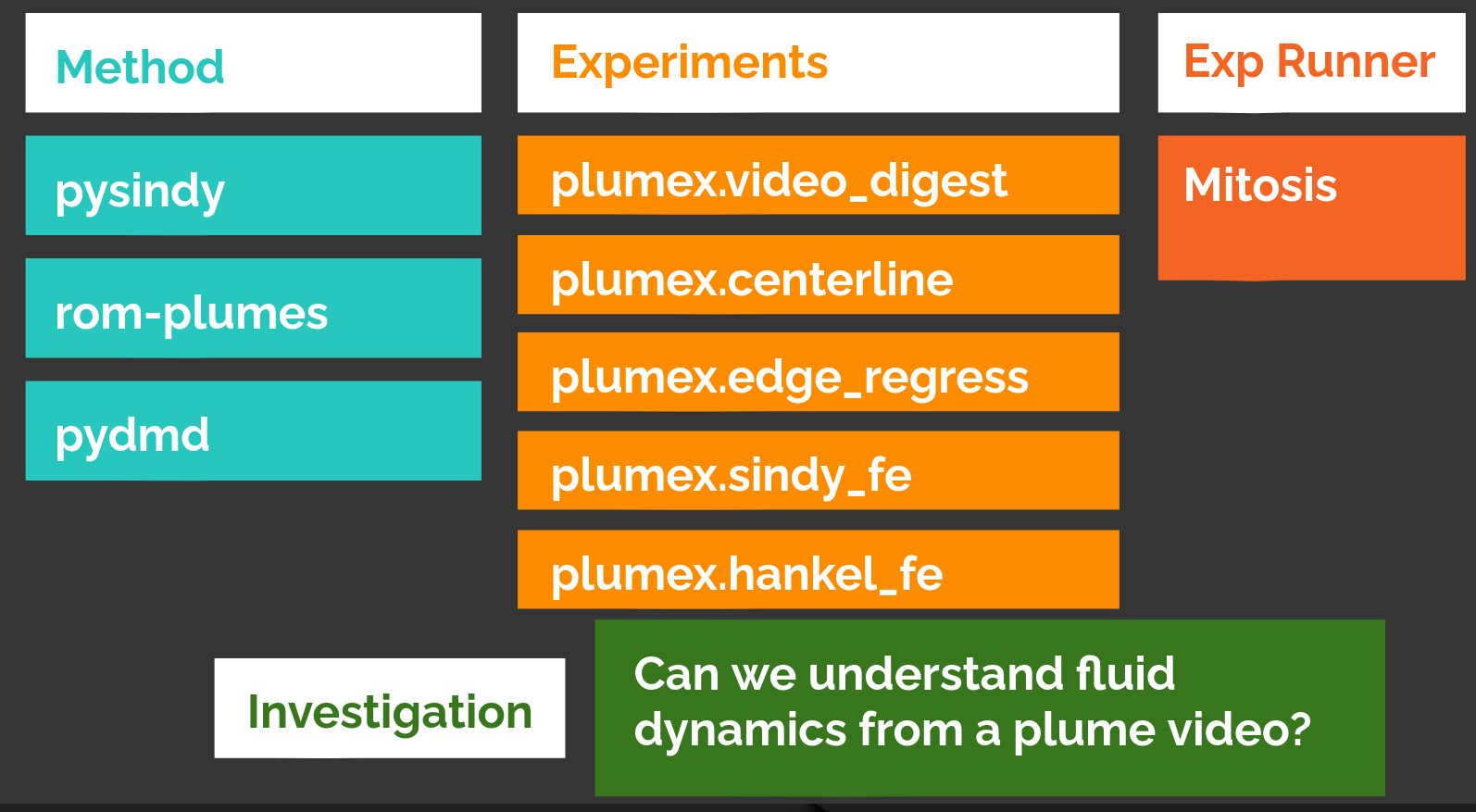
A depiction of the project organization for the plumes project. This consists of a package to process plume videos into various reduced-order models, a package to run experiments to evaluate different parameterizations of those reduced order models, a copy of mitosis, and a project that represents the investigations in the paper and presentation, specifying the particular choices of parameters to investigate. This layout was used for several hundred invocations of experiments as we improved and debugged the experiments and identified the best parameterizations.
Since the investigation often evolves alongside the methods and experiments, I prefer to leave those as separate distribution packages. To make dependency updates simpler, I incorporate them as git submodules within the overall investigation repository. Alternatively, you can include method and experiment as parts of the investigation repository and distribution package, but kept as different import packages. Regardless, this separation of concerns helps force decisions about what choices are generally applicable and which are specific to the data in an investigation. Long term, this helps people further innovate the methods without stopping to wonder why decisions were made that don't truly belong with the code they're editing.
When working on a remote server with collaborators:
- Everyone works on a different clone of the of the project folder, but the trials folder is symlinked to a shared directory.
- An HTTP server runs in the background within that shared trial directory. This allows users to use their browser to view trial artifacts, as well as linking to them in a meetings notes google doc. (e.g. nohup python -m http.server 8080 &)
- My .ssh/config handles port forwarding to allow me to browse the SSH server's local HTTP development server using eg. LocalForward 8000 localhost:8080
Lookup dictionary
The main reason to specify the arguments of interest as a dictionary is standardize the human names among collaborators. Otherwise, these arguments occur in a variety of default values and spread throughout a notebooks definition. However, an unintended benefit is how concise the the git diff of a dictionary can be.
The diff becomes a form of communication, and tools that generate communication from work products are a huge aid to collaboration. To take a case in point, it's easy to see from the example below what my kinds of experiments my colleague below was working on:
When experiments are factored well, its easy to see what parameters other people are investigating.
Dependency inversion
The relationship between the experiment runner and experiments is the perfect situation to use dependency inversion. That is, in order for the runner to be project independent, it cannot depend upon the experiments. Yet in order for the experiments to be project independent, albeit to a lesser degree, they can't import the runner.
I admit I fumble a bit explaining dependency inversion, but this example is the essence of it for me: The existence of the higher level facility (runner) does not depend upon the lower-level library, and the lower level library does not depend upon the higher level runner.
The Python facility for entry points is a good example of how this works. Normally, the lower-level library has responsibility for registering the entry point. Our separation of concerns, however, puts that responsibility on the investigation. The investigation's pyproject.toml includes a tool.mitosis table for naming different experiments and associating their lookup dictionaries. This has the added benefit of allowing references to short names of experiments but still disambiguating any name conflicts. E.g. I can use experiments written by other folks such as grad_project.utils.dmd_fit_eval and lab_utils.dmd.dmd_fit_eval, but refer them locally in my investigation as jess_dmd, and lab_dmd.
OBTW: The use of ex.sh
It's also useful to keep a file ex.sh in the investigation repository to track the current "official" experiments. This allows you to move experiments to a new machine in case the funding agency is yoinking back those expensive GPU servers and you get left off the email notification.